Poster #P29
Identification of critical internal coordinates to sample reaction paths of enzymatic reactions
Modeling enzymatic reaction pathways is a challenging and also computationally demanding task. The numerous degrees of freedom in an enzymatic system, out of which many can be relevant for the reaction and its energetic profile, at least indirectly, render the notion of “the reaction mechanism”, read a single reaction pathway, naïve. One promising approach to tackle this issue is by using the transition network [1].
Our system of interest, Carboxypeptidase A (CPA), which contains a divalent zinc ion in its active site, is an important exopeptidase secreted by the pancreas for digesting intake proteins in the metabolism cycle. It catalyzes the elimination of the C-terminal amino acid via hydrolysis, with a preference for residues with hydrophobic side chains. Thus we want to develop a method to automatically sample the reaction pathways and find the most probable route by an automated choice of degrees of freedom for the sampling.
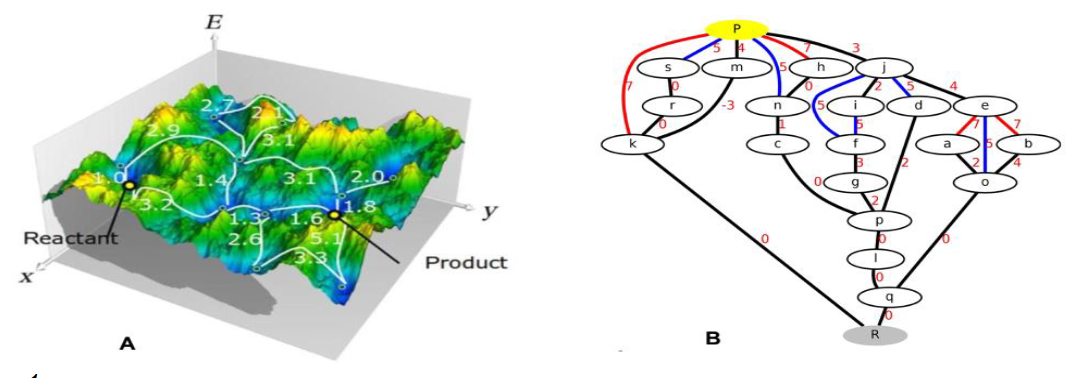
Figure 1. A) Schematic energy landscape with valleys (blue) with low and mountains (yellow/orange) with high energy, respectively. Yellow points mark end states and green dots are intermediate states. White connections with transition barriers indicate a variety of possible pathways. Figure is taken from ref 2. B) Example of a transition network with nodes (reactant, products and intermediates), weighted edges(paths joining 2 intermediates with the corresponding transition energy).
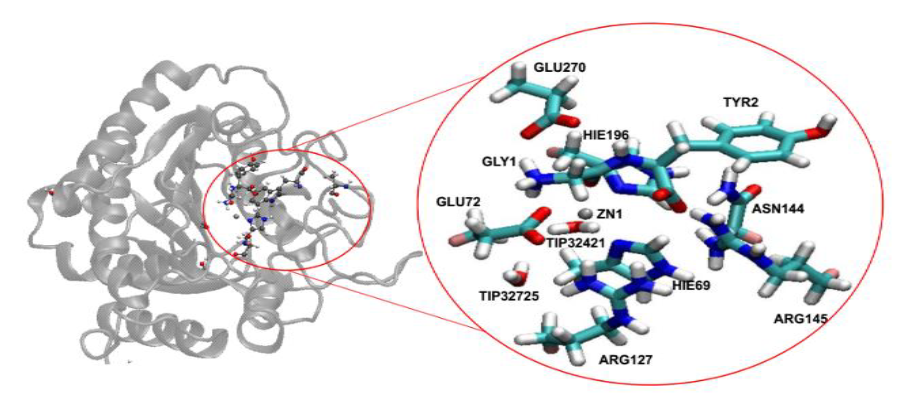
Figure 2. Left) Crystal structure of Carboxypeptidase A. Right) The corresponding active site.
- F. Noe, D. Krachtus, J. C. Smith and S. Fischer, J. Chem. Theory Comput. 2006, 2, 840-857.
- P. Imhof, Computational Approaches for Studying Enzyme Mechanism Part B 2006, 578, 11.
Epee Ndongue Jules César
- Computer-Chemistry-Center and Friedrich-Alexander-Universität Erlangen-Nürnberg