Poster #P33
Exploration of the Chemical Reaction Space of Cezanne-2 through MD and QM/MM
Cezanne-2 (Cez2) is a deubiquitinating enzyme which is involved in the regulation of ubiquitin-driven cellular signalling and a potential target for the development of drugs against cancer and neurological disorders [1,2]. It selectively targets Lys11-linked polyubiquitin chains. Its exact catalytic mechanism, however, is not fully resolved yet. Understanding the latter may help in the rational design of selective inhibitors.
In this work, we use extensive molecular dynamics (MD) simulations in explicit water to study the interaction of di-ubiquitin substrate with Cez2 and thus get molecular insight into Cez2 catalysis. Since protein crystallography does not allow the assignment of protonation states, we explored the structural dynamics of Cez2 in different charge/protonation states of the catalytic Cys/His dyad (Fig. 1; [3]): i) both His367 (singly protonated at Nε) and Cys210 (protonated) with neutral charge, and ii) His367 (doubly protonated) positively charged and Cys210 (deprotonated) negatively charged. We modelled both the apo enzyme and the di-ubiquitin Cez2 complex. Four independent MD simulations were performed for each system, using OpenMM and the CHARMM36 force field.
The simulations provide information about the structural arrangements of the catalytic core of Cez2 in pre- and post-reactive enzymatic states. Particular reactive spatial configurations of di-ubiquitin are identified which are susceptible to hydrolysis by Cez2 (i.e., productive configurations), thus providing very detailed information on the chemical space transformation during catalysis. The reliability of the productive configurations were verified by QM/MM (B3LYP/TZVP//CHARMM36) optimizations.
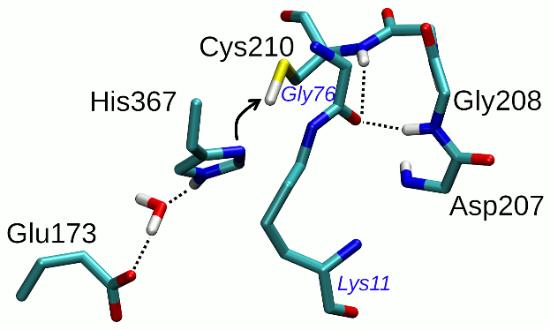
Figure 1. Cez2 di-ubiquitin complex optimized by QM/MM. For clarity, only relevant parts of both the catalytic core of Cez2 (labels in black) and di-ubiquitin (labels in blue) are shown.
- Su, S., Chen, J., Jiang, Y., et. al., Adv. Sci. 2021, 8, 2004846.
- Suzuki, H., Inaba, M., Yamada, M., et. al., Am. J. Med. Genet. A. 2021, 185, 1182-1186.
- Ilter, M., Schulze-Niemand, E., et. al., J. Chem. Inf. Model. 2023, 63, 2084-2094.
Andrés M. Escorcia
- Max Planck Institute for Dynamics of Complex Technical Systems, Magdeburg (DE)